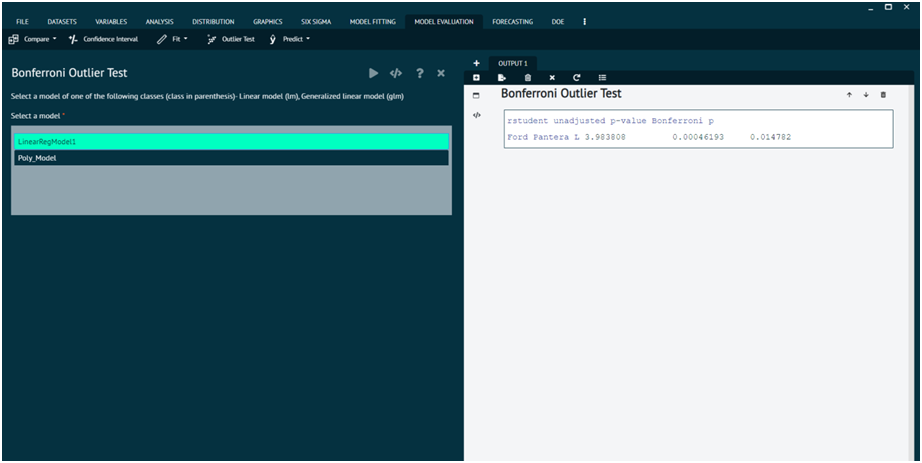
Outlier Test
Bonferroni Outlier Test
For a linear model, p-values reported use the t distribution with degrees of freedom one less than the residual df for the model. For a generalized linear model, p-values are based on the standard-normal distribution. The Bonferroni adjustment multiplies the usual two-sided p-value by the number of observations.
This function reports the Bonferroni p-values for testing each observation in turn to be a mean-shift outlier, based Studentized residuals in linear (t-tests), generalized linear models (normal tests), and linear mixed models.
Arguments
- model
an lm, glm, or lmerMod model object; the "lmerMod" method calls the "lm" method and can take the same arguments.
- cutoff
observations with Bonferroni p-values exceeding cutoff are not reported, unless no observations are nominated, in which case the one with the largest Studentized residual is reported.
- n.max
maximum number of observations to report (default, 10).
- order
report Studenized residuals in descending order of magnitude? (default, TRUE).
- labels
an optional vector of observation names.
- ...
arguments passed down to methods functions.
- x
outlierTest object.
- digits
number of digits for reported p-values.