Factor analysis
Factor analysis is a statistical technique used to identify and analyze underlying factors or latent variables that explain the observed correlations among a set of variables.
The goal of factor analysis is to reduce the dimensionality of the data by identifying a smaller number of latent factors that explain the observed correlations among variables. This can simplify the interpretation of complex datasets and help identify underlying patterns or structures.
Factor analysis can be conducted using various statistical software packages, and BioStat Prime utilized R packages to conduct factor analysis. BioStat Prime brings forth 2 ways of factor analysis, viz.
Factor
Principal Component Analysis.
Principal Component Analysis (PCA) and Factor Analysis (FA) are both techniques used in multivariate analysis to uncover patterns and relationships in high-dimensional data. However, they serve different purposes, and it's important to distinguish between them.
While PCA and Factor Analysis share similarities, their primary objectives differ. PCA is primarily a variance-driven technique for dimensionality reduction, while Factor Analysis is a model-based technique for understanding the underlying structure of the data in terms of latent factors. The choice between them depends on the research question and the nature of the data.
Factor Analysis
Perform maximum-likelihood factor analysis on a covariance matrix or data matrix and generates a screeplot.
To analyse Factor Analysis in BioStat Prime user must follow the steps as given.
- Steps
Load the dataset -> Click on the analysis tab in main menu -> Select factor analysis tab -> Select Factor -> Once the dialog appears choose the items to be included -> Specify no. of factors to be extracted, user can also save factors and take a scree plot -> Execute the dialog.
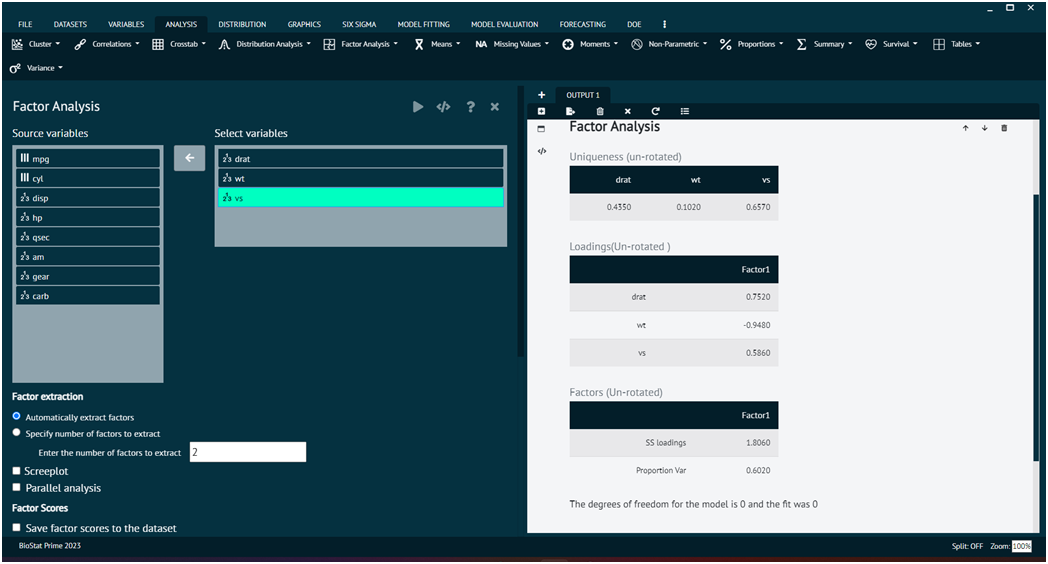
For further information the user can explore model tuning and model evaluation options for the same.
The following are the different type argument:
- vars
One or more numeric variables to extract factors from.
- autoextraction
Automatically determine the number factors or extract specific numbers of factors.
- screeplot
If TRUE generates a screeplot.
- rotation
determine the type of rotation and takes one of the values (none, quartimax, geominT, varimax, oblimin, simplimax, promax, geominQ and bentlerQ)
- saveScores
saves the factor scores in the dataset
- dataset
The dataset from which the 'vars' have been picked.
Principal Component Analysis
Performs a principal components analysis on the given numeric data matrix and returns the results as an object of class princomp.
To analyse Principal Component Analysis in BioStat Prime user must follow the steps as given.
- Steps
Load the dataset -> Click on the analysis tab in main menu -> Select factor analysis tab -> Select Principal Component Analysis -> Once the dialog appears choose the items to be included -> Specify no. of factors to be extracted, user can also save factors and take a scree plot -> Execute the dialog.
For further information the user can explore model tuning and model evaluation options for the same.
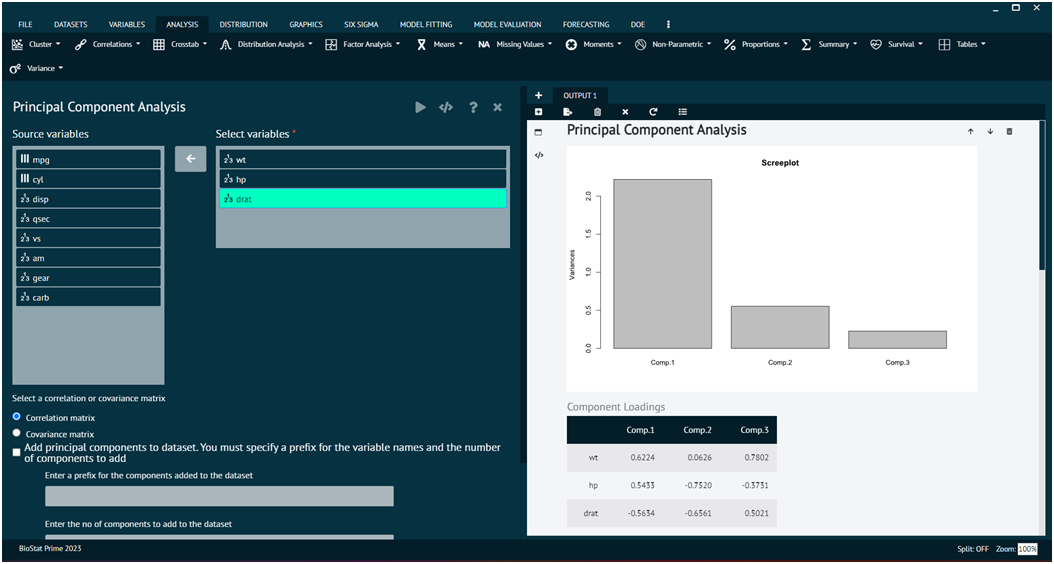
The following are the different type argument:
- vars
The variables in a character vector to extract components from
- cor
A boolean that specifies whether the calculation should use a correlation or covariance matrix
- componentsToRetain
A numeric that Specifies the number of components to retain in the dataset. A new variable is created in the dataset for each component invoked
- generateScreeplot
Generates a screeplot
- prefixForComponents
Prefix to use when saving the components to a dataset
- dataset
The name of the dataset as a string